Understanding the molecular and cellular processes that cause dementia is one of the most important challenges in neuroscience. SUMOylation is a post-translational protein modification that has been strongly implicated in neurodegenerative diseases. To investigate SUMOylation in dementia we profiled the expression of key SUMOylation pathway proteins in post mortem brain tissue from Alzheimer’s Disease (AD) and Down’s Syndrome (DS) patients. As expected, both AD and DS tissue displayed massively increased levels of phosphorylated tau compared to age- and sex-matched controls. Surprisingly, there were no changes in the levels of the E1 and E2 enzymes required for protein SUMOylation, or in levels of the deSUMOylating enzyme SENP1. There was, however, a marked decrease in the SUMO-2/3-specific deSUMOylating enzyme SENP3 in DS. There were also increased levels of SUMO-1 conjugated proteins in DS, but not in AD tissue. While these results do not exclude roles for SUMOylation in AD, they demonstrate clear differences in the profile of SUMOylation and in the expression of deSUMOylating enzymes between AD and DS brain.
Introduction
Dementia is defined as a condition that results in memory disorders, personality changes, and impaired reasoning. Alzheimer’s Disease (AD) is the most common cause of dementia and affects over 35 million people worldwide (Reitz et al. 2011). It is already one of the leading causes of death in the developed world and the number of new cases is predicted to double in the next 30 years, putting an ever-increasing strain on medical and social care resources.
Primary histopathological characteristics of AD are the accumulation of neurofibrillary tangles composed of a hyperphosphorylated form of the microtubule-associated protein tau, and extracellular plaques comprising aggregated beta amyloid (Aβ) peptides, which are cleavage products produced from the amyloid precursor protein (APP) (Reitz et al. 2011). To generate Aβ, APP is sequentially cleaved by β-site APP cleaving enzyme (BACE), followed by γ-secretase, which cleaves APP within its transmembrane domain to yield Aβ peptides with lengths between 38 and 43 amino acids which can aggregate into extracellular oligomers and accumulate to form senile plaques (Lee et al. 2013). Aβ40 is the most common Aβ peptide accounting for ~90% of secreted Aβ, but Aβ42 has the highest propensity for aggregation (Hilbich et al. 1991). Together, these aggregates lead to a devastating decrease in the number of neurons in multiple brain regions, including the hippocampus, where up to 60-70% of neurons are lost by the late-stage of the disease (West et al. 1994) and in the cortex, resulting in the cognitive decline and dementia characteristic of AD.
Down’s Syndrome (DS) is a congenital disorder caused by an extra copy (trisomy) of chromosome 21, which results in intellectual impairment and physical abnormalities. DS is the most common genetic cause of intellectual disability (Wiseman et al. 2009). Improved medical treatment and social inclusion has increased the life expectancy of people with DS to more than 55 years (Glasson et al. 2002). However, by the age of 40 most DS sufferers develop amyloid plaques and neurofibrillary tangles, and by the age of 60, ~60% have dementia (Wiseman et al. 2009). This has been attributed, at least in part, to overexpression of the chromosome 21 gene products APP (Portelius et al. 2014) and the kinase DYRK1A (Wiseman et al. 2009), which mediates hyperphosphorylation of tau.
Small ubiquitin-like modifier (SUMO) proteins are ~10kD proteins that can be covalently conjugated to lysine residues in target proteins to modify their function. In humans, there are three SUMO paralogues (SUMO-1-3); SUMO-2 and SUMO-3 are identical except for three residues, but they share only ~50% sequence identity with SUMO-1 (Wilkinson and Henley 2010, Henley et al. 2014). Protein SUMOylation by both SUMO-1 and SUMO-2/3 has been strongly implicated in a wide range of neuropathologies including AD, Parkinson’s Disease and Huntington’s Disease (McMillan et al. 2011, Sarge and Park-Sarge 2011, Nistico et al. 2014) although many of the target proteins have not yet been determined. Nonetheless, recent studies have suggested that increased levels of SUMOylation, and of specific substrates in particular, may play important neuroprotective roles in cell stress responses (Guo et al. 2013, Guo and Henley 2014). SUMO proteins are conjugated to target proteins by an enzymatic cascade consisting of three steps: SUMO is first activated for conjugation in an ATP-dependent manner by an E1 enzyme, a heterodimer consisting of SUMO conjugating enzyme subunit 1 and 2 (SAE1 and SAE2) in humans, before it is passed to the catalytic cysteine of the sole SUMO conjugating E2 enzyme Ubc9 (Ubiquitin-conjugating 9). Ubc9, either alone or in combination with an E3 enzyme, then catalyses SUMO transfer to a lysine residue in a target protein (Geiss-Friedlander and Melchior 2007, Wilkinson and Henley 2010). Importantly, SUMOylation is reversible and can be removed from substrates by the actions of a number of SUMO-specific proteases, the best-characterised of which are the SENP family of cysteine proteases. In humans, there are 6 SENPs (SENP1, 2, 3, 5, 6 and 7), which differ in their preference for cleaving SUMO-1 versus SUMO-2/3 and in their subcellular localisation (Hickey et al. 2012). For example, SENP1 exhibits broad specificity for SUMO-1 and SUMO-2/3, whereas SENP3 preferentially deconjugates SUMO-2/3 from target proteins (Guo and Henley 2014). Thus, the SUMOylation status of individual target proteins is the result of a delicate balance between E1/E2-dependent conjugation, and SUMO-protease-dependent removal. The levels and activity of these enzymes is therefore crucial in determining the cellular SUMO proteome.
Both tau and APP are reported to be SUMO substrates. Tau is SUMOylated at Lys340 (Dorval and Fraser 2007, Lee et al. 2013) and SUMO-1 immunoreactivity co-localizes with phospho-tau aggregates in transgenic AD model mice (Takahashi et al. 2008). APP was identified as a putative SUMO substrate in a systematic search for SUMO target proteins (Gocke et al. 2005). Subsequently, it has been shown that Lys587 and Lys595, which are immediately N-terminally adjacent to the BACE cleavage site, are covalently modified by SUMO-1 and SUMO-2. Preventing SUMOylation of these lysine residues was reported to increase Aβ aggregation, whereas promoting APP SUMOylation by expressing the SUMO E2 conjugating enzyme Ubc9 decreased Aβ aggregation in HeLa cells (Zhang and Sarge 2008). Consistent with this, decreased Aβ aggregation has also been observed upon overexpression of SUMO-3 in human embryonic kidney and neuroblastoma cells (Li et al. 2003). However, in apparent contrast, SUMO-3 overexpression has been shown to cause a significant increase in toxic Aβ40 and Aβ42 secretion and, surprisingly, this effect was independent of the ability of SUMO-3 to conjugate to target proteins (Dorval and Fraser 2007). Although inconclusive, these studies are of particular interest because the genes for both APP and SUMO-3 are located on chromosome 21.
Intriguingly, it has recently been reported that SUMOylation is required for normal synaptic plasticity since inhibition of SUMOylation with a dominant negative Ubc9 peptide blocks LTP (Jaafari et al. 2013, Lee et al. 2014). Furthermore, enhancing protein SUMOylation can overcome deficits in LTP induced by application of Aβ42, and global protein SUMOylation by SUMO-2/3 was suggested to be dysregulated in the brains of AD patients and in the Tg2576 transgenic mouse model of AD (Lee et al. 2014).
A core aim of this project was to explore if levels of the SUMO machinery enzymes are altered in brain samples from patients suffering from clinically diagnosed and categorised late stage dementia. In particular, we were interested if there was any detectable imbalance between levels of selected SUMO conjugating enzymes versus levels of SUMO deconjugating enzymes in samples from people with dementia compared to age- and sex-matched controls.
Methods
Brain samples: Post mortem tissue from the Human Tissue Authority licensed South West Dementia Brain Bank was used with approval from the Research Ethics Committee. Anterior prefrontal cortex (Brodmann Area 10, BA10) was used from 20 AD patients (ages 64–95, mean=78.7 ± 8.6 SD; Post mortem delay (PMD)=7.5–73 hours, mean=38.6 ± 20.8 SD) and 20 age- and sex-matched control cases (ages 68–97, mean=85.1 ± 8.6 SD; PMD=5.5–76 hours, mean=42.4 ± 3.2 SD). BA10 samples from 9 DS patients were analysed (ages 48-67, mean=59.9 ± 6.5 SD; PMD=16-76 hours, mean=39.2 ± 20.8 SD) and 9 age-matched controls (ages 43-72, mean=62 ± 8.9; PMD=4-66 hours, mean=25.2 ± 20 SD).
Sample preparation: 100mg of tissue was homogenized in 445μl of ice-cold Lysis Buffer (25 mM Hepes pH 7.4, 150 mM NaCl, 1 x protease inhibitor, 20 mM N-ethylmaleimide (NEM, Sigma-Aldrich)), transferred to a 1.5 ml Eppendorf tube and solubilised by adding 50μl 10% Triton X-100 and 5μl 10% SDS (for a final concentration of 1% or 0.1%, respectively), sonicated briefly and gently agitated for 1 hour at 4°C. Insoluble material was pelleted by centrifugation at 16,000g for 15 minutes at 4°C. The supernatant was removed and centrifuged again at 16,000g for 30 minutes at 4°C, after which the supernatant was transferred to a fresh 1.5ml Eppendorf tube. The pellets of both centrifugation steps were retained and stored at -80°C. The protein concentration in the supernatant was determined by Bradford assay, divided into 50 μl aliquots and stored at -80°C. Prior to SDS-PAGE analysis samples were thawed and treated with 1μl of Benzonase Nuclease for 30 min at 4°C to digest all nucleic acids prior to addition of 2x SDS-PAGE Sample Buffer and boiling the samples for 5 minutes at 95°C or incubation overnight at room temperature (RT).
SDS-PAGE and Western blotting: Proteins were resolved using commercially prepared 26 well 4–20% gradient SDS-PAGE gels (BioRad) according to the manufacturer’s instructions. Molecular weight was determined using PageRuler Prestained Protein Ladder (Thermo Scientific). 15μl (27.75μg protein) was loaded in each lane and following electrophoresis the proteins were transferred onto PVDF membrane and blocked in 5% non-fat milk powder in TBS-T. Proteins were labelled by incubation with primary antibodies for 1 hour at RT and secondary antibodies for 45 minutes at RT. Following extensive washes immunolabelled protein was detected using Pierce ECL Western Blotting Substrate and image capture on a LI-COR Odyssey Fc.
Primary antibodies used were: Rabbit anti-APP (C-terminal, Sigma, 1:1000); Mouse anti-Tau (Millipore, 1:1000); Mouse anti-Phospho-Tau (Cell Signalling Technology, 1:1000); Mouse anti-SUMO-1 (Santa Cruz Biotechnology, 1:500); Sheep anti-SUMO-2/3 (a gift from Prof. Ron Hay (University of Dundee), 1:1000); Mouse anti-SENP1 (Santa Cruz Biotechnology, 1:500); Rabbit anti-SENP3 (Cell Signalling Technology, 1:1000); Goat anti-AOS1 (SAE1) (Santa Cruz Biotechnology, 1:250); Rabbit anti-Ubc9, (Santa Cruz Biotechnology, 1:500); Mouse anti-β-tubulin type III (Sigma, 1:10,000).
Analysis and quantification: The density of immunoreactive bands of interest determined by a LI-COR scanner were normalised to type III β-tubulin levels. For SUMO-1 and SUMO-2/3 all bands in the lane were measured whereas for individual proteins of interest the specific immunoreactive bands were quantified. Data were analysed using a student’s t-test and presented as mean ± SEM with the mean of the control group set to 100%. Significance level was set at p<0.05.
Results and Discussion
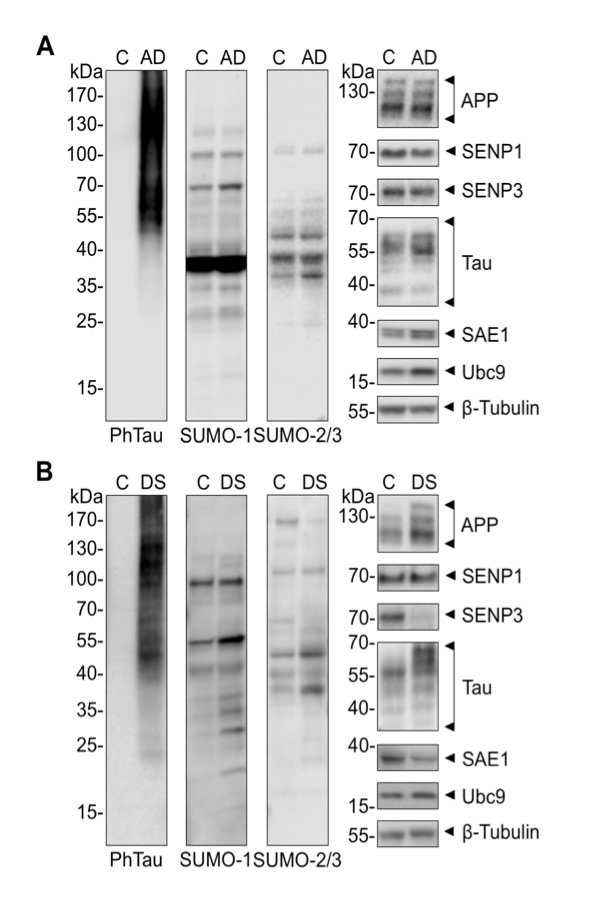
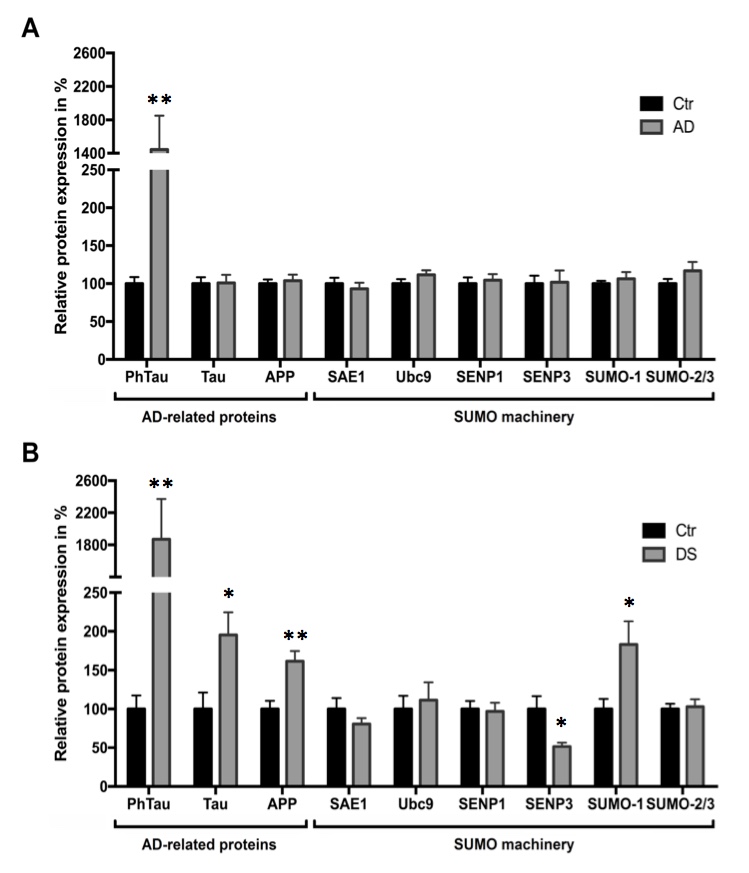
We obtained brain samples from the BA10 region of the anterior prefrontal cortex from 20 AD patients, 9 DS patients, and corresponding age- and sex-matched controls. Tissue homogenates were prepared from these samples, prior to analysis by Western blotting. We first validated the disease phenotype in post mortem samples by measuring levels of phosphorylated tau, which is a well-established marker for AD and DS pathology (Wiseman et al. 2009, Di Domenico et al. 2013). In agreement with previous reports, we observed a >10-fold increase in phospho-tau in both AD and DS patients compared to age-matched controls (p=0.0022 and p=0.0021, respectively).
There was also a significant increase in total levels of tau in DS (p=0.02), but not in AD samples. Consistent with the fact that APP is encoded on trisomic chromosome 21, APP was increased in DS samples (p=0.002) but unchanged in AD samples (Fig. 1, 2).
Figure 1. Representative Western blots for each of the proteins analysed in BA10 homogenates from AD (A), DS (B) and control samples. (A) Brain homogenates of BA10 from 20 age- and sex-matched control patients were subjected to Western blot analysis using anti-phospho-Tau, -tau, -APP, -SUMO-1, -SUMO-2/3, -SAE1, -Ubc9, -SENP1, -SENP3 and –β-tubulin III antibodies. (B) Post mortem brain tissue from 9 DS and 9 control patients were analysed by Western blotting using anti-phospho-Tau, -tau, -APP, -SUMO-1, -SUMO-2/3, -SAE1, -Ubc9, -SENP1, -SENP3 and –β-tubulin III antibodies.
Contrary to our initial expectations, total levels of the E1 SUMO activating enzyme subunit SAE1 and the E2 conjugating enzyme Ubc9 were unchanged in diseased versus control BA10 samples (Fig. 1, 2). Notably, while there were no changes in the deSUMOylating enzymes SENP1 and SENP3 in AD, there was a significant decrease in SENP3 (p=0.01) in DS tissue.
Furthermore, no significant differences in the total levels of free SUMO-1 or SUMO-2/3, or in the overall patterns of proteins conjugated to SUMO were detected in AD samples. Intriguingly, however, there was a significant increase in SUMO-1 (p=0.02), but not SUMO-2/3, conjugates in DS samples. At first sight this is unexpected since SUMO-3 is encoded on chromosome 21 (Hattori et al. 2000), and SUMOylation by SUMO-2/3 has been reported to be increased in DS (Gardiner 2006). However, this study only examined one control and one DS case, in contrast to the 9 control versus 9 DS analysed here. Our data suggest that SUMO-3 trisomy does not influence the steady-state level of SUMO-2/3 modification, however, this may potentially be explained by recent data suggesting that SUMO-2 is the predominant SUMO-2/3 isoform, at least in mice (Wang et al. 2014). As a result, the increased SUMO-3 may not be observed in DS since it still remains a relatively minor proportion of the total SUMO-2/3 protein present. We were unable to examine levels of SUMO-4 or the recently reported SUMO-5 paralogues (Liang et al. 2016) due to the lack of antibodies specific to these proteins. However, since the importance of SUMO-4 as a genuine ubiquitin-like modifier protein has been questioned (Owerbach et al. 2005) and SUMO-5 expression is not detectable in brain (Liang et al. 2016), it seems unlikely that these proteins play a major role in the pathology of AD or DS.
Figure 2. Histogram demonstrating normalised levels of the indicated proteins in BA10 samples taken post mortem from AD (A), DS (B) and control samples. (A) Quantification of the expression levels of the proteins of interest after normalising to the neuron-specific loading control β-tubulin III. Phosphorylated tau is significantly increased (p=0.0022) in BA10 of AD compared to control patients. No significant changes in tau, APP or SUMO machinery proteins (SAE1, Ubc9, SENP1, SENP3, SUMO-1, SUMO-2/3) were observed (p>0.05, n=20, unpaired two-tailed student’s t-test). Data presented as mean + SEM with the mean of the control group set to 100%.(B) Proteins of interest were normalised to corresponding β-tubulin III levels and quantification showed a significant increase of phospho-Tau (p=0.0021), tau (p=0.02), APP (p=0.002) and SUMO-1 (p=0.02) in DS compared to control samples, whereas SENP3 was found to be significantly decreased (p=0.01).
Our initial hypothesis was that, because SUMOylation has emerged as an important regulator of synaptic and neuronal function and dysfunction, the SUMOylation machinery would be altered in AD and DS brain (Guo and Henley 2014, Henley et al. 2014). To our knowledge this is the first investigation of protein SUMOylation in DS and we observed a significant increase in SUMO-1 conjugation and a loss of the SUMO-2/3 deconjugating enzyme SENP3. At first sight this is surprising since the loss of SENP3 would be expected to correlate with an increase in SUMO-2/3 conjugation rather than SUMO-1. However DS is a complex disease and we attribute the change in the steady-state level of SUMO-1 conjugation to other mechanisms. In addition, while we did not observe global changes in SUMO-2/3 conjugation, we did observe changes in the level of some SUMO-2/3 modified substrates at ~170kD (p=0.03, Fig. 1). Moreover, the loss of SENP3 will alter the SUMOylation status of individual substrates that may not be apparent by Western blotting for global SUMOylation levels. Relevant to this, we have previously shown that, during ischemia, SENP3 is rapidly degraded, and that this promotes cell survival upon reperfusion by promoting SUMOylation of the SENP3 substrate Drp1 (Guo et al. 2013). Taken together our data suggest that, similar to ischemia, the cell stress associated with dementia in DS may lead to a loss of SENP3, potentially as part of a neuroprotective stress response.
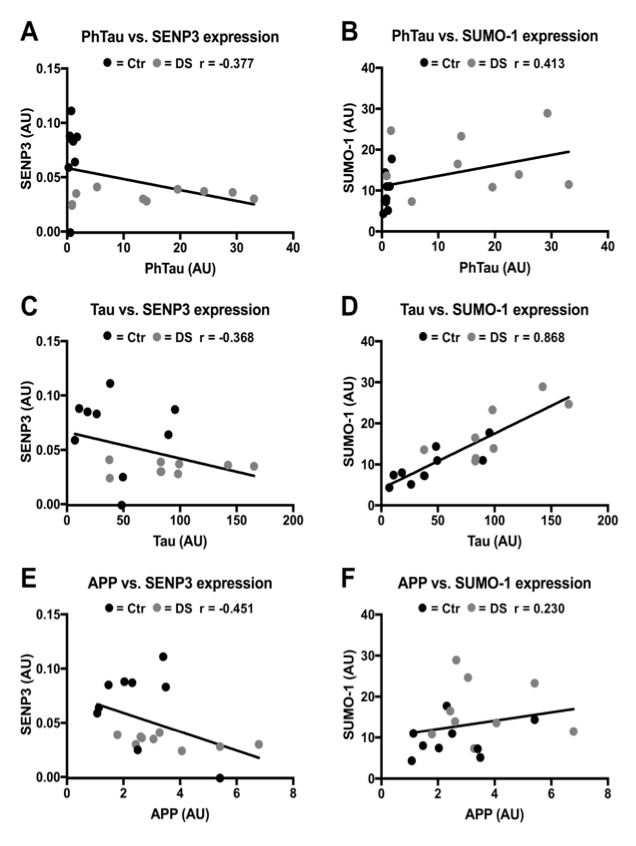
To further examine the link between phospho-Tau, total Tau and APP markers of disease pathology and the changes observed in DS brain, we examined correlation between these sets of proteins in the data from the control and DS patients (Fig. 3). Interestingly, we observed a significant positive correlation between levels of Tau expression and SUMO-1 conjugation. However, it is important to note that this does not necessarily indicate a direct causal link between increased Tau levels and SUMO-1-ylation in DS, and further work will be required to investigate this link. Nonetheless, in AD, where Tau levels were unchanged, we did not observe any changes in total SUMO-1-ylation, consistent with this possible relationship.
Figure 3. Correlation between SUMO pathway proteins with significantly altered expression in DS and markers of disease pathology. (A) Scatterplot showing no correlation between PhTau and SENP3 expression (r=-0.377, p=0.12, n=18). (B) Scatterplot showing no correlation between PhTau and SUMO-1 expression (r=0.413, p=0.09, n=18). (C) Scatterplot showing no correlation between Tau and SENP3 expression (r=-0.363, p=0.13, n=18).(D) Scatterplot showing a significant positive correlation between Tau and SUMO-1 expression (r=0.868, p<0.0001, n=18). (E) Scatterplot showing no correlation between APP and SENP3 expression (r=-0.451, p=0.06, n=18). (F) Scatterplot showing no correlation between APP and SUMO-1 expression (r=0.230, p=0.36, n=18).
In contrast to DS, neither SUMO substrate conjugation nor SUMO conjugating and deconjugating enzymes were changed in AD. Indeed, these results are broadly in line with previous data on SUMOylation and selected SUMO enzyme levels in transgenic mice overexpressing APP (McMillan et al. 2011).
Unlike DS, which has a readily identifiable genetic cause, AD is a heterogeneous group of diseases with a diverse range of potential underlying factors. Consistent with this diversity, we observed large variation between the individual AD samples reflecting the heterogeneity of the disease. Thus, it is likely that only very large and robust changes associated with common pathways in all forms of AD will be detected in post mortem tissue.
These results are supported by a recent report showing that there is no change in Ubc9 or SUMO-1 levels in AD (Lee et al. 2014). However, that study observed a significant decrease in protein SUMOylation by SUMO-2/3 in AD hippocampus. There are several possible reasons for these apparent differences. The sample size in that study was relatively small, comprising hippocampal samples from only 6 controls and 6 AD samples and they used β-actin as a loading control (Lee et al. 2014). However, it is important to note that our study examined cortex rather than hippocampus and used a much larger sample size (20 AD and 20 control brains), which may partially explain the differences in our observations.
It is important to emphasize that although we observed no significant differences in total levels of the subset of SUMO pathway conjugating and deconjugating enzymes examined in AD samples, the lack of change in expression does not necessarily indicate that the SUMOylation pathway is unaffected in this disease. We are assessing enzymes, and it is the conjugating and catalytic activity that is critical in determining the levels of SUMOylation and deSUMOylation of substrate proteins. Thus, it remains possible that the activity or localisation of the proteins and enzymes involved might be drastically changed even if their expression is unaltered. Furthermore, in terms of levels of substrate SUMOylation, we can only assess the overall patterns and total levels of substrate modification by SUMO paralogues. In this study we do not address modification of individual substrates, and it needs to be kept in mind that the SUMOylation status of key individual substrates may be dramatically altered, even in the absence of gross changes in global SUMOylation levels.
In summary, we compared post mortem brain samples from patients who died with AD or DS to age- and sex-matched control samples from patients who died of unrelated causes. Our hypothesis was that, because of the emerging key role of post-translational protein modification by SUMO in regulating synaptic function and health, the SUMO pathway might be profoundly affected in AD and/or DS pathology. Our results suggest that although there are some differences in protein SUMOylation and enzyme levels in DS, in AD in particular there is little evidence for major alterations in the levels of the enzymes that regulate protein SUMOylation. We note, however, that our data do not exclude key changes in the SUMOylation status of specific substrate proteins in the onset and progression of the synaptic deficiencies associated with DS and AD.
| Attachment | Size |
|---|---|
| 693.98 KB |
We would like to thank the South West Dementia Brain Bank (SWDBB) for providing brain tissue for this study. The SWDBB is supported by BRACE (Bristol Research into Alzheimer's and Care of the Elderly), Brains for Dementia Research and the Medical Research Council. We also thank Prof. Ron Hay (University of Dundee) for the gift of the sheep polyclonal SUMO-2/3 antibody. CSB is a BRACE-funded PhD student and MJH is an Alzheimer’s Society-funded PhD student. This work was supported by research funding from the MRC, BRACE and the Alzheimer’s Society.
Di Domenico, F., R. Coccia, A. Cocciolo, M. P. Murphy, G. Cenini, E. Head, D. A. Butterfield, A. Giorgi, M. E. Schinina, C. Mancuso, C. Cini and M. Perluigi (2013). "Impairment of proteostasis network in Down syndrome prior to the development of Alzheimer's disease neuropathology: redox proteomics analysis of human brain." Biochim Biophys Acta 1832(8): 1249-1259.
Dorval, V. and P. E. Fraser (2007). "SUMO on the road to neurodegeneration." Biochimica Et Biophysica Acta-Molecular Cell Research 1773(6): 694-706.
Gardiner, K. (2006). "Transcriptional dysregulation in Down syndrome: predictions for altered protein complex stoichiometries and post-translational modifications, and consequences for learning/behavior genes ELK, CREB, and the estrogen and glucocorticoid receptors." Behav Genet 36(3): 439-453.
Geiss-Friedlander, R. and F. Melchior (2007). "Concepts in sumoylation: a decade on." Nat Rev Mol Cell Biol 8(12): 947-956.
Glasson, E. J., S. G. Sullivan, R. Hussain, B. A. Petterson, P. D. Montgomery and A. H. Bittles (2002). "The changing survival profile of people with Down's syndrome: implications for genetic counselling." Clin Genet 62(5): 390-393.
Gocke, C. B., H. Yu and J. Kang (2005). "Systematic identification and analysis of mammalian small ubiquitin-like modifier substrates." J Biol Chem 280(6): 5004-5012.
Guo, C. and J. M. Henley (2014). "Wrestling with stress: roles of protein SUMOylation and deSUMOylation in cell stress response." IUBMB Life 66(2): 71-77.
Guo, C., K. L. Hildick, J. Luo, L. Dearden, K. A. Wilkinson and J. M. Henley (2013). "SENP3-mediated deSUMOylation of dynamin-related protein 1 promotes cell death following ischaemia." EMBO J 32(11): 1514-1528.
Hattori, M., A. Fujiyama, T. D. Taylor, H. Watanabe, T. Yada, H. S. Park, A. Toyoda, K. Ishii, Y. Totoki, D. K. Choi, Y. Groner, E. Soeda, M. Ohki, T. Takagi, Y. Sakaki, S. Taudien, K. Blechschmidt, A. Polley, U. Menzel, J. Delabar, K. Kumpf, R. Lehmann, D. Patterson, K. Reichwald, A. Rump, M. Schillhabel, A. Schudy, W. Zimmermann, A. Rosenthal, J. Kudoh, K. Schibuya, K. Kawasaki, S. Asakawa, A. Shintani, T. Sasaki, K. Nagamine, S. Mitsuyama, S. E. Antonarakis, S. Minoshima, N. Shimizu, G. Nordsiek, K. Hornischer, P. Brant, M. Scharfe, O. Schon, A. Desario, J. Reichelt, G. Kauer, H. Blocker, J. Ramser, A. Beck, S. Klages, S. Hennig, L. Riesselmann, E. Dagand, T. Haaf, S. Wehrmeyer, K. Borzym, K. Gardiner, D. Nizetic, F. Francis, H. Lehrach, R. Reinhardt, M. L. Yaspo, m. Chromosome and c. sequencing (2000). "The DNA sequence of human chromosome 21." Nature 405(6784): 311-319.
Henley, J. M., T. J. Craig and K. A. Wilkinson (2014). "Neuronal SUMOylation: mechanisms, physiology, and roles in neuronal dysfunction." Physiol Rev 94(4): 1249-1285.
Hickey, C. M., N. R. Wilson and M. Hochstrasser (2012). "Function and regulation of SUMO proteases." Nat Rev Mol Cell Biol 13(12): 755-766.
Hilbich, C., B. Kisters-Woike, J. Reed, C. L. Masters and K. Beyreuther (1991). "Aggregation and secondary structure of synthetic amyloid beta A4 peptides of Alzheimer's disease." J Mol Biol 218(1): 149-163.
Jaafari, N., F. A. Konopacki, T. F. Owen, S. Kantamneni, P. Rubin, T. J. Craig, K. A. Wilkinson and J. M. Henley (2013). "SUMOylation is required for glycine-induced increases in AMPA receptor surface expression (ChemLTP) in hippocampal neurons." PLoS One 8(1): e52345.
Lee, L., E. Dale, A. Staniszewski, H. Zhang, F. Saeed, M. Sakurai, M. Fa, I. Orozco, F. Michelassi, N. Akpan, H. Lehrer and O. Arancio (2014). "Regulation of synaptic plasticity and cognition by SUMO in normal physiology and Alzheimer's disease." Sci Rep 4: 7190.
Lee, L., M. Sakurai, S. Matsuzaki, O. Arancio and P. Fraser (2013). "SUMO and Alzheimer's disease." Neuromolecular Med 15(4): 720-736.
Li, Y., H. Wang, S. Wang, D. Quon, Y. W. Liu and B. Cordell (2003). "Positive and negative regulation of APP amyloidogenesis by sumoylation." Proc Natl Acad Sci U S A 100(1): 259-264.
Liang, Y. C., C. C. Lee, Y. L. Yao, C. C. Lai, M. L. Schmitz and W. M. Yang (2016). "SUMO5, a Novel Poly-SUMO Isoform, Regulates PML Nuclear Bodies." Sci Rep 6: 26509.
McMillan, L. E., J. T. Brown, J. M. Henley and H. Cimarosti (2011). "Profiles of SUMO and ubiquitin conjugation in an Alzheimer's disease model." Neurosci Lett 502(3): 201-208.
Nistico, R., C. Ferraina, V. Marconi, F. Blandini, L. Negri, J. Egebjerg and M. Feligioni (2014). "Age-related changes of protein SUMOylation balance in the AbetaPP Tg2576 mouse model of Alzheimer's disease." Front Pharmacol 5: 63.
Owerbach, D., E. M. McKay, E. T. Yeh, K. H. Gabbay and K. M. Bohren (2005). "A proline-90 residue unique to SUMO-4 prevents maturation and sumoylation." Biochem Biophys Res Commun 337(2): 517-520.
Portelius, E., M. Holtta, H. Soininen, M. Bjerke, H. Zetterberg, A. Westerlund, S. K. Herukka, K. Blennow and N. Mattsson (2014). "Altered cerebrospinal fluid levels of amyloid beta and amyloid precursor-like protein 1 peptides in Down's syndrome." Neuromolecular Med 16(2): 510-516.
Reitz, C., C. Brayne and R. Mayeux (2011). "Epidemiology of Alzheimer disease." Nat Rev Neurol 7(3): 137-152.
Sarge, K. D. and O. K. Park-Sarge (2011). "SUMO and its role in human diseases." Int Rev Cell Mol Biol 288: 167-183.
Takahashi, K., M. Ishida, H. Komano and H. Takahashi (2008). "SUMO-1 immunoreactivity co-localizes with phospho-Tau in APP transgenic mice but not in mutant Tau transgenic mice." Neurosci Lett 441(1): 90-93.
Wang, L., C. Wansleeben, S. Zhao, P. Miao, W. Paschen and W. Yang (2014). "SUMO2 is essential while SUMO3 is dispensable for mouse embryonic development." EMBO Rep 15(8): 878-885.
West, M. J., P. D. Coleman, D. G. Flood and J. C. Troncoso (1994). "Differences in the pattern of hippocampal neuronal loss in normal ageing and Alzheimer's disease." Lancet 344(8925): 769-772.
Wilkinson, K. A. and J. M. Henley (2010). "Mechanisms, regulation and consequences of protein SUMOylation." Biochem J 428(2): 133-145.
Wiseman, F. K., K. A. Alford, V. L. Tybulewicz and E. M. Fisher (2009). "Down syndrome--recent progress and future prospects." Hum Mol Genet 18(R1): R75-83.
Zhang, Y. Q. and K. D. Sarge (2008). "Sumoylation of amyloid precursor protein negatively regulates Abeta aggregate levels." Biochem Biophys Res Commun 374(4): 673-678.